Decoding Deep-Sea Fish Genomes: Revealing How Vertebrates Conquered High-Pressure Abyssal Environments
Decoding Deep-Sea Fish Genomes: Revealing How Vertebrates Conquered High-Pressure Abyssal Environments
On March 6, 2025, a major research achievement on deep-sea fishes—led by Researcher Shunping He and jointly conducted with the CAS Institute of Deep-Sea Science and Engineering, the Institute of Hydrobiology, Northwestern Polytechnical University, and other institutions—was published in Cell under the title “Evolution and genetic adaptation of fishes to the deep sea.”
Developed under the Global TREnD Program and based on deep-sea specimens collected using China’s independently developed deep-diving technologies, the study delivers the first comprehensive, multi-dimensional breakthrough spanning genomics, physiology, ecology, and evolutionary biology.
National Deep-Sea Equipment: Driving Scientific Exploration Into the Hadal Realm
The research relied on the mothership “TAN SUO YlHAO” (exploration l) and “TAN SUO ER HAO” (exploration ll), and the manned submersible “SHEN HAI YONG SH!”(Deep-SeaWarrior), “FEN DOU ZHE" (Striver), and the remotely operatedvehicle (ROV)“TIAN YA”(Skyline).
Across multiple expeditions spanning the Mariana Trench, Yap Trench, Diamantina and Wallaby–Zenith trenches, the Southwest Indian Ocean hydrothermal fields, the Philippine Basin, and the South China Sea, the team sampled nearly the entire depth range of deep-sea fishes (1,218–7,730 m), obtaining 11 species across six major deep-sea lineages, including six hadal (>6,000 m) species.
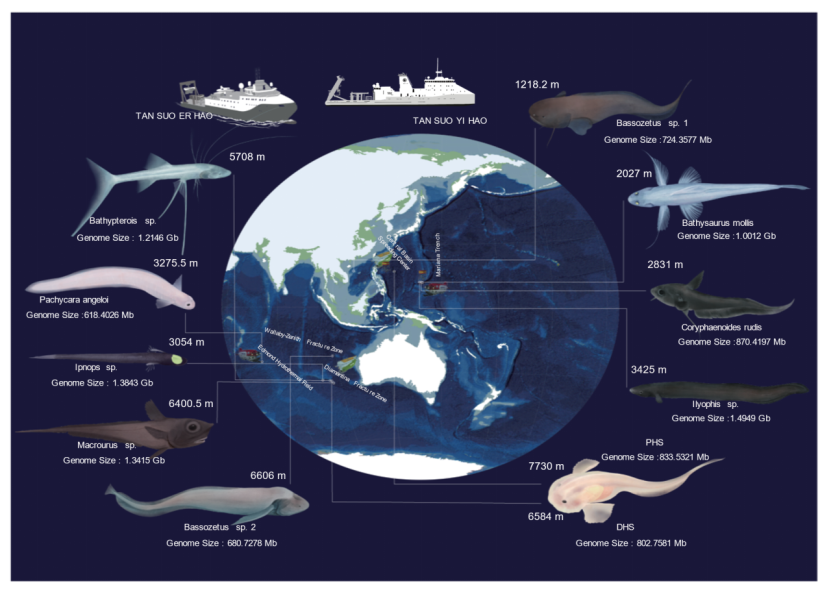
Fig 1. Sampling information and morphological characteristics of 11 deep-sea species.
Major Findings
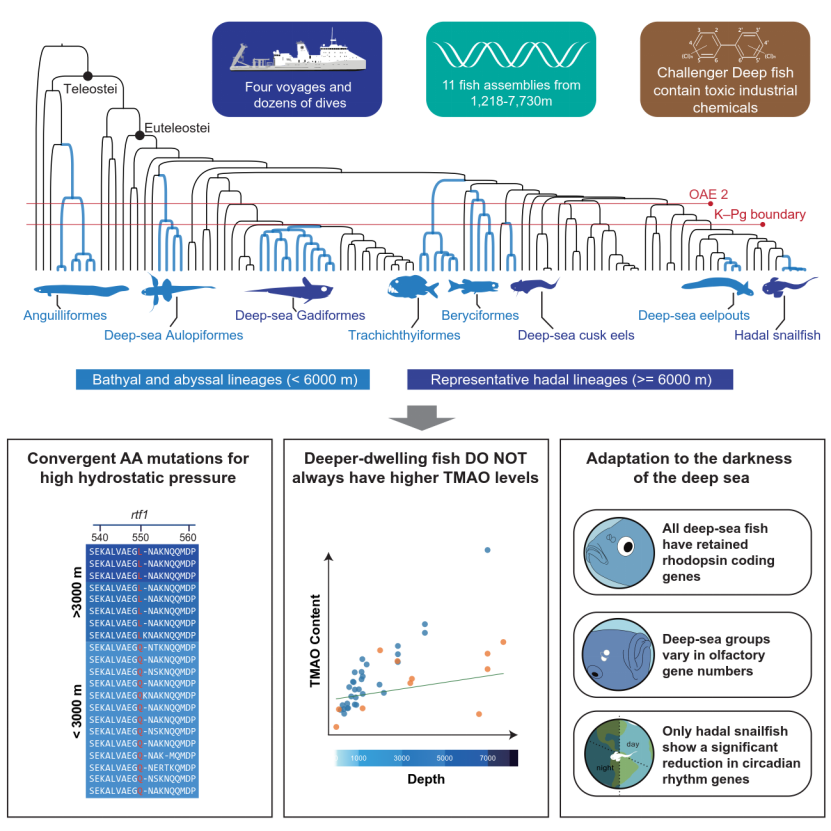
Fig 2. Graphical abstract
The study analyzed the genomes of 11 deep-sea fish species collected across the Western Pacific and Central Indian Ocean at depths from 1,218 to 7,730 m, reconstructing a detailed evolutionary tree of deep-sea fishes and tracing the history of vertebrate colonization of the deep sea.
Results show that most living deep-sea fishes entered the deep ocean only after the Cretaceous–Paleogene mass extinction (~66 Ma), while a few ancient lineages began adapting to deep-sea environments over 100 million years ago, surviving multiple global extinction events. The genomes also exhibit lower mutation rates, higher repeat content, and multi-level adaptive changes to perpetual darkness.
The team quantified TMAO levels in fishes across different depths and found that while species living between 0–6,000 m follow the known trend of increasing TMAO with depth, this pattern disappears beyond 6,000 m. This demonstrates that TMAO alone cannot explain vertebrate survival under extreme high pressure in the hadal zone, suggesting the existence of more sophisticated molecular mechanisms.
A groundbreaking finding in the study is the discovery of a highly conserved amino acid substitution—Q550L in the rtf1 gene—present in all deep-sea fishes inhabiting below 3,000 m. Functional experiments show that this mutation significantly alters transcriptional efficiency, revealing that transcriptional regulation may play a key role in adaptation to extreme hydrostatic pressure. This discovery opens a new avenue for exploring vertebrate high-pressure adaptation and highlights the importance of transcriptional control in deep-sea survival.
The team detected extremely high concentrations of polychlorinated biphenyls (PCBs) in the livers of Mariana and Philippine Trench snailfishes. This alarming finding demonstrates that anthropogenic pollutants have already accumulated in Earth’s deepest ecosystems, underscoring the urgent need for deep-sea environmental protection and pollution monitoring.
The study also revealed lineage-specific adaptive strategies. Different deep-sea fishes have evolved distinct solutions to cope with extreme pressure, cold, and darkness. Meanwhile, analyses of metabolites—such as fatty acids, amino acids, and heavy metals—along with proteomic data, provided deeper insights into the physiological adaptations of representative species. Population genomic analyses of hadal snailfish showed that deep-ocean currents may facilitate cross-trench gene flow, revealing unexpected connectivity between geographically distant trench populations.
The project was supported by the CAS Strategic Priority Research Program (Category B), the CAS International Partnership Program, the National Key R&D Program, the National Natural Science Foundation of China, Hainan Provincial Science and Technology Program, and the Global TREnD initiative.
Here is the link to the paper you referenced: https://doi.org/10.1016/j.cell.2025.01.002